Objects in R
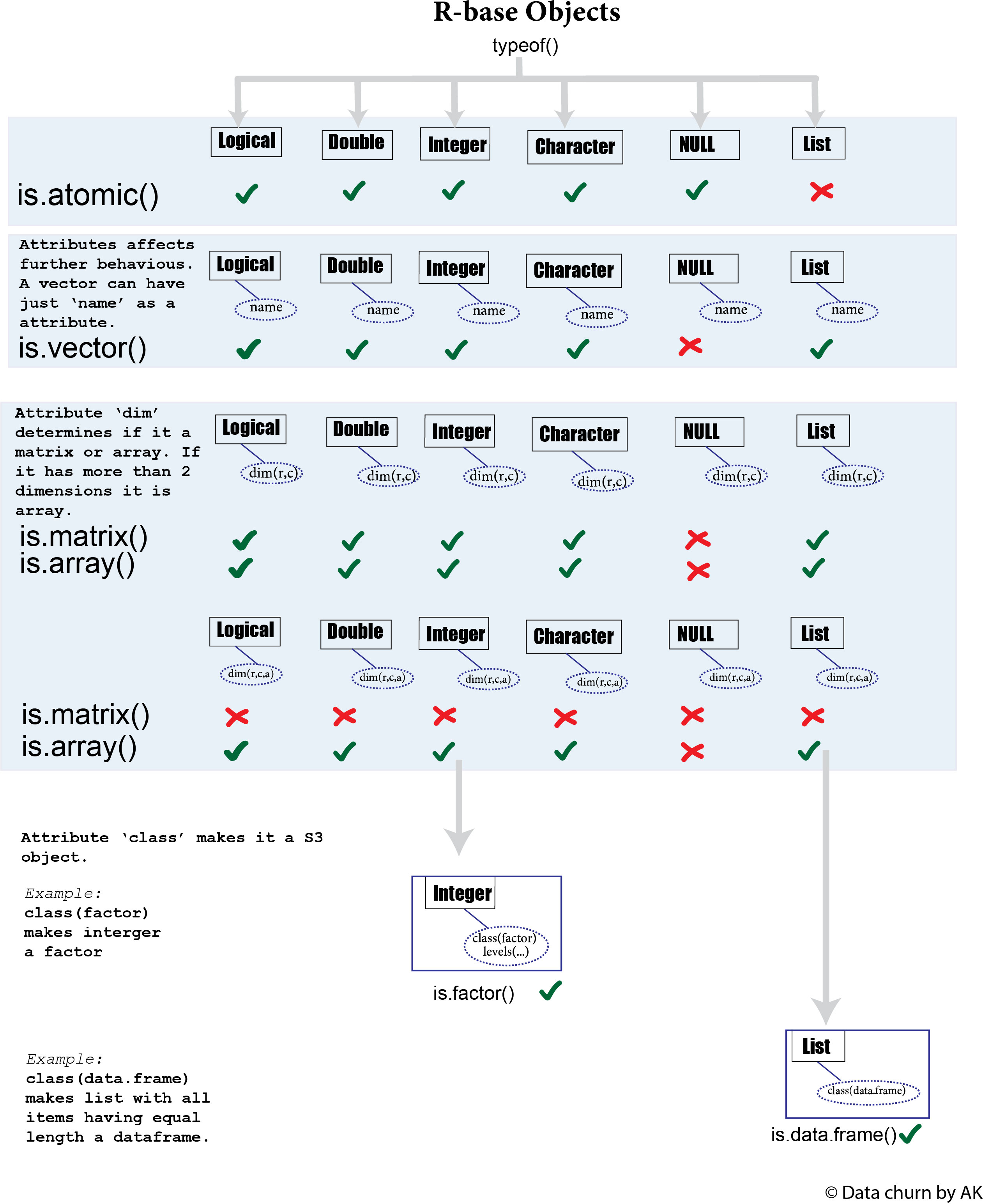
R by design is a functional programming language hence the original objects designed to represent variables, functions, environments and various language components are different than the objects used in true Object Oriented Programs. True OOP-like objects S3, S4 were added in different time during the evolution of R and build upon the base R objects. The function typeof() gives the base-type of any objects (including S3, S4) in R. Another function mode() gives similar output but groups integers and double as numeric. There are 25 different types of base R objects but six are worth mentioning as they are used to represent variable/data set and most commonly encountered by R users.
- Logical: TRUE or FALSE / 1 or 0
- Double: Numbers with decimal values
- Integer: Numbers without decimal values
- Character: strings
- NULL : No value
- List: List of any combination of above five types
All the other data types we make in R like matrix, array, data.frame, tibble etc. are derived almost entirely from these 6 object types. But before going to that it is important clarify some terms.
1. Atomic object : If an object cannot hold its own object type inside itself it is called atomic object. For example: a vector of logical, double, integer, character and NULL. The values in atomic objects are stored in unique memory locations. Can be represented like the figure below:
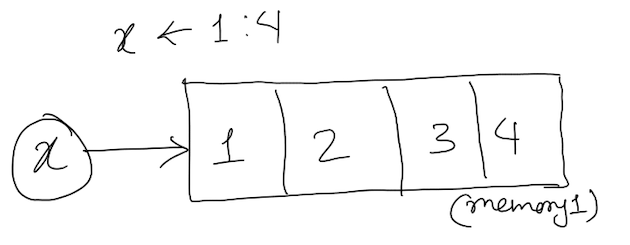
The function is.atomic() is always true for logical, double, integer, character and NULL objects even if they have higher dimensions or name attribute.
NB: For each of the atomic object type these is a special value “NA”, which represent missing values and missing values can be identified by using function is.na().
a_logic <- as.logical(sample(0:1, 12, replace=T)) #create a logical of length 12
a_double <- rnorm(12) #create a double of length 12
an_integer <- 1:12 #create an integer
a_char <- letters[1:12] #create a character
a_null<- NULL
##See what type of object they are:
typeof(a_logic)
## [1] "logical"
# typeof(a_double)
# typeof(an_integer)
# typeof(a_char)
# typeof(a_null)
##See if they are atomic or not
is.atomic(a_logic)
## [1] TRUE
# is.atomic(a_double)
# is.atomic(an_integer)
# is.atomic(a_char)
# is.atomic(a_null)
##They will remain as atomic even after adding higher dimensions
dim(an_integer) <- c(3,4)
is.atomic(an_integer) #Still true
## [1] TRUE
2. Recursive object: These are the objects which can hold its own object type within itself. For example list can contain another list inside itself. It is represented by following figure.
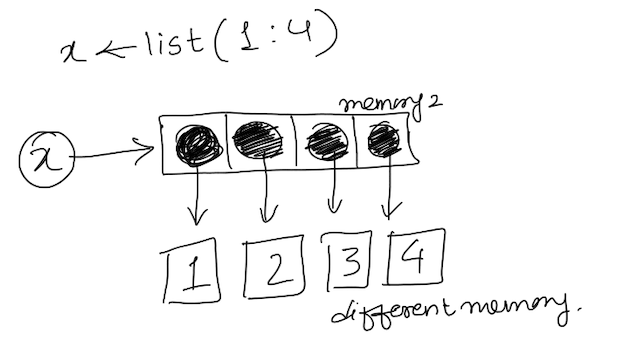
The opposite of function is.atomic() is is.recursive().
a_list <- list(1:9, 2:4, "a", 3:5)
is.atomic(a_list) #FALSE
## [1] FALSE
is.recursive(a_list) #TRUE
## [1] TRUE
Attributes
Attributes are the metadata associated with each R-objects. This metadata further controls how an object is processed inside R and how we perceive an object to be. One of the simplest attribute is name which gives a name to each value in a vector.
Vector
Any base R objects can be a vector (including a list) as long as it has no any attributes or single attribute name. Use the function attributes() to get all the attributes associated with a object and function attr() to get a specific attribute. Once other attributes like dim or class are added it is no more called a vector and the function is.vector() will be FALSE.
attributes(a_double) # the vector we created has no attributes, gives NULL
## NULL
is.vector(a_list) ##A simple list is also a vector
## [1] TRUE
names(a_list) <- c("list1", "list2","list3", "list4")
a_list #print named list
## $list1
## [1] 1 2 3 4 5 6 7 8 9
##
## $list2
## [1] 2 3 4
##
## $list3
## [1] "a"
##
## $list4
## [1] 3 4 5
is.vector(a_list) #it is still a vector
## [1] TRUE
Matrix
Adding another attribute dim makes a vector (including a list although less common in practice) into a array. Matrix is a special case of array where we have only two dimensions (rows and columns). Matrix is treated as special because it is in the form in which many data are represented (a table with rows and columns.). Therefore there are some function in R that are specific to a matrix like (rownames(), colnames(), nrow(), ncol(), rbind(), cbind(), t()) which also applies to a data.frame in R. One major difference between matrix and data.frame is base data type of matrix is atomic type (character, integer, double …) but data.frame is stored as a list. Because of the nature of atomic objects, matrix build from atomic objects should have same datatype in all columns.
dim(a_double) <- c(3,4) #this makes a matrix out of variable a_double with 3 rows and 4 columns and filled columns first
a_double
## [,1] [,2] [,3] [,4]
## [1,] 0.9822688 -0.07954649 1.62053229 -1.46717395
## [2,] -0.7089867 -0.10584293 -1.47000813 1.81499311
## [3,] -0.3611035 1.41759061 -0.03876332 0.04078724
is.vector(a_double) #It is no more a vector
## [1] FALSE
is.atomic(a_double) #But is still a atomic
## [1] TRUE
dim(a_list) <- c(2,2) # a matrix of list can also be made but it won't look like a data.frame
a_list
## [,1] [,2]
## [1,] integer,9 "a"
## [2,] integer,3 integer,3
Array
When a object has 2 or more dimension as attribute it is called a array.
dim(a_double) <- c(2,3,2)
a_double # The third dimension is shown by breaking the table, it can be regarded as two 2X3 tables
## , , 1
##
## [,1] [,2] [,3]
## [1,] 0.9822688 -0.36110347 -0.1058429
## [2,] -0.7089867 -0.07954649 1.4175906
##
## , , 2
##
## [,1] [,2] [,3]
## [1,] 1.620532 -0.03876332 1.81499311
## [2,] -1.470008 -1.46717395 0.04078724
Class and S3 object
Attribute class is very important because it powers the whole OOP in R. Once a R base object has class attribute it is called a S3 object. Commonly used S3 objects are factors, dates, data and time, data.frame, and tibble. Because of this very lenient criteria, it is possible to assign any object to class which it should not belong to. For example we can assign a object of type double as data.frame. R will not stop us from doing this but since double will not be compatible with functions related to data.frame we will eventually run us into an error. Therefore we should always use a constructor function provided for a class to make a class instead of simply assigning a class. Any object with class attribute can be identified by function is.object().
a_double <- rnorm(12)
dim(a_double) <- c(6,2)
a_double #looks like a data.frame
## [,1] [,2]
## [1,] 0.10532213 -0.725692115
## [2,] 0.63655714 -0.003428998
## [3,] 1.82306620 1.132239738
## [4,] 0.11533008 -0.055686044
## [5,] -0.05375573 1.180911669
## [6,] 0.22075057 -0.016582626
class(a_double) <- "data.frame" ##assign a new class to object a_double
attributes(a_double) # now we see a_double has class attribute which is data.frame
## $dim
## [1] 6 2
##
## $class
## [1] "data.frame"
class(a_double) #it says a_double has a class = data.frame
## [1] "data.frame"
a_double[1,1] #hit an error trying to use a data.frame function
## NULL
## <0 rows> (or 0-length row.names)
##proper way
a_double <- rnorm(12)
dim(a_double) <- c(6,2)
df <- data.frame(a_double) #use the constructor function.
df
## X1 X2
## 1 0.4021199 0.57215428
## 2 -0.9181690 0.71649773
## 3 -0.2014391 1.98107538
## 4 -1.5345385 2.00999835
## 5 0.3284315 -0.64234315
## 6 0.6641933 -0.01521207
class(df) ##now it is actually a data.frame
## [1] "data.frame"
Factor
Factor is one of the most used and also one of the most confused S3 object in R. It is build on top of the base R object integer and must have two attributes:
- class: must be “factor”
- levels: must be atomic vector of unique values in factor.
Because it is a S3 object the generic function print will behave specific to factor and prints the factors at the position indicated by the integers in actual object.
a_char <- c("a","b","c","a","c","b","d")
typeof(a_char)
## [1] "character"
print(a_char)
## [1] "a" "b" "c" "a" "c" "b" "d"
as.numeric(a_char) ##No numeric is produced
## Warning: NAs introduced by coercion
## [1] NA NA NA NA NA NA NA
a_factor <- as.factor(a_char)
print(a_factor) #now it is a factor so we see same character
## [1] a b c a c b d
## Levels: a b c d
typeof(a_factor) #under the hood it is an integer
## [1] "integer"
attributes(a_factor)
## $levels
## [1] "a" "b" "c" "d"
##
## $class
## [1] "factor"
as.numeric(a_factor) #gives the underlying integer
## [1] 1 2 3 1 3 2 4
data.frame
It is a S3 object which is build upon a list. Individual columns are stored like a individual items in a list. So data.frame is actually a list where all the items in list must have equal length.
S4 object
Because S3 objects are easy to incorporate errors another OOP type object called S4 was build which is extensively used by bioconductor community. Most of these S4 objects are build on base R object type list.
library(edgeR)
## Loading required package: limma
a<-DGEList()
## Warning in min(lib.size): no non-missing arguments to min; returning Inf
## Warning in min(norm.factors): no non-missing arguments to min; returning Inf
typeof(a) ##under the hood it is a list
## [1] "list"
class(a) ##gives class "DGEList"
## [1] "DGEList"
## attr(,"package")
## [1] "edgeR"
attributes(a) #shows all the attributes
## $class
## [1] "DGEList"
## attr(,"package")
## [1] "edgeR"
##
## $names
## [1] "counts" "samples"